TF-Finder
Abstract
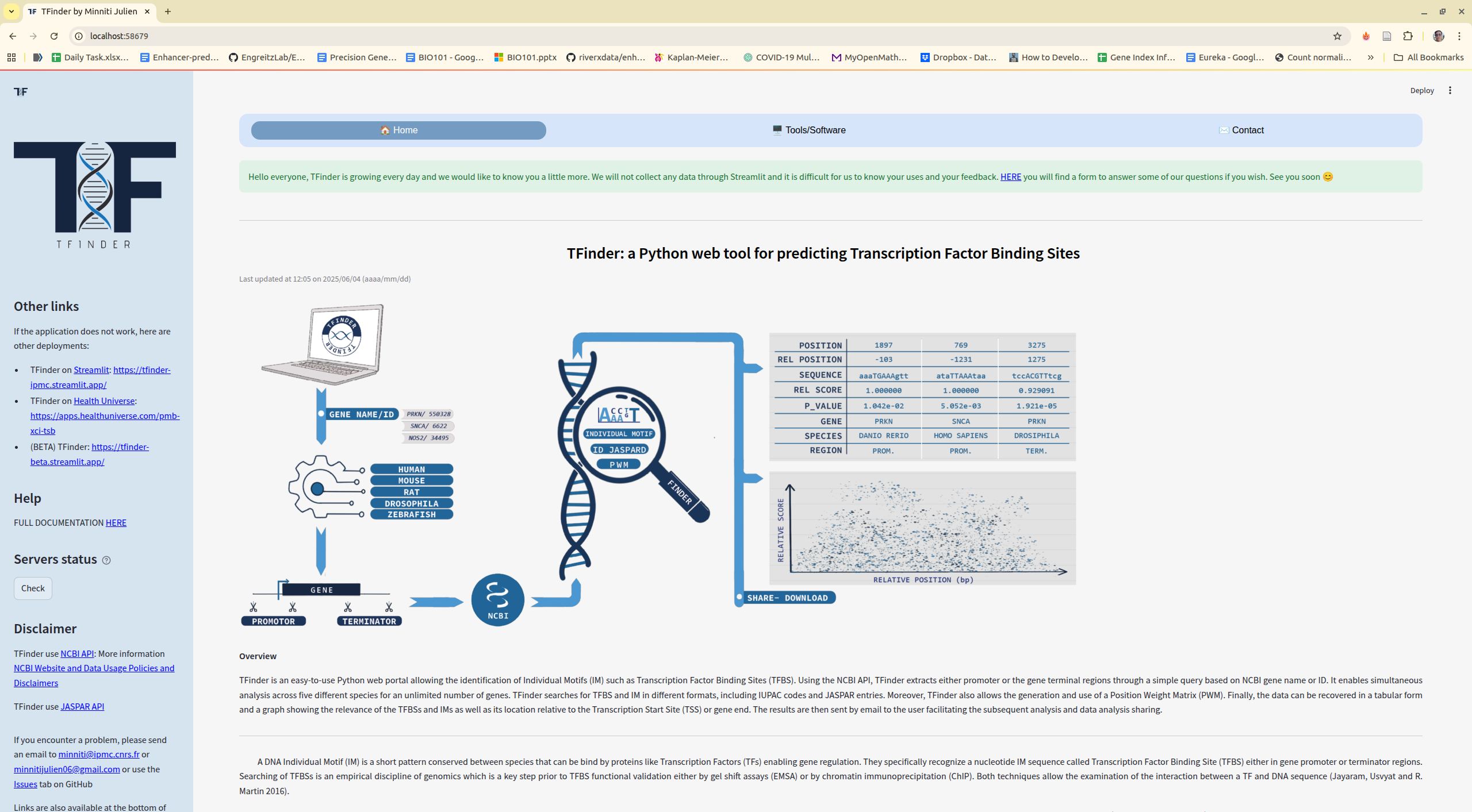
Transcription is a key cell process that consists of synthesizing several copies of RNA from a gene DNA sequence. This process is highly regulated and closely linked to the ability of transcription factors to bind specifically to DNA. TFinder is an easy-to-use Python web portal allowing the identification of Individual Motifs (IM) such as Transcription Factor Binding Sites (TFBS). Using the NCBI API, TFinder extracts either promoter or gene terminal regulatory regions, through a simple query of NCBI gene name or ID. It enables simultaneous analysis across five different species for an unlimited number of genes. TFinder searches for Individual Motifs in different formats, including IUPAC codes and JASPAR entries. Moreover, TFinder also allows de novo generations of a Position Weight Matrix (PWM) and the use of already established PWM. Finally, the data are provided in a tabular and a graph format showing the relevance and the P-value of the Individual Motifs found as well as their location relative to the Transcription Start Site (TSS) or the terminal region of the gene. The results are then sent by email to users facilitating the subsequent data analysis and sharing. TFinder is written in Python and freely available on GitHub under the MIT license: https://github.com/Jumitti/TFinder. It can be accessed as a web application implemented in Streamlit at https://tfinder-ipmc.streamlit.app. Resources are available on Streamlit “Resources” tab. TFINDER strength is that it relies on an all-in-one intuitive tool allowing users inexperienced with bioinformatics tools to retrieve gene regulatory regions sequences in multiple species and to search for individual motifs in a huge number of genes.
Citation
@article{minniti2025tfinder,
title={TFinder: A Python Web Tool for Predicting Transcription Factor Binding Sites},
author={Minniti, Julien and Checler, Frederic and Duplan, Eric and da Costa, Cristine Alves},
journal={Journal of Molecular Biology},
volume={437},
number={3},
pages={168921},
year={2025},
publisher={Elsevier}
}
RIVER integration
- It is the public tools, new configuration is added to run on RIVER
- Available at: https://github.com/riverxdata/bioinfor-sw-TFinder/
For existing bioinformatics tools with web interfaces, simply add a river/main.sh file. Recommended approach: use pixi for environment management or singularity for containerized tools.
TFFinder is a Streamlit application that demonstrates the adaptation process.
Step 1: Fork the Repository
Fork TFFinder if you're not the owner. See GitHub forking documentation.
Step 2: Clone the Repository
git clone git@github.com:riverxdata/bioinfor-sw-TFinder.git
cd bioinfor-sw-TFinder
Step 3: Add Testing Infrastructure
git clone git@github.com:riverxdata/web-template-analysis.git
cp -r web-template-analysis/tests tests
rm -rf web-template-analysis
Step 4: Create the RIVER Integration
Create river/main.sh with the following content:
cd $RIVER_HOME/jobs/$job_id/analysis
ENV=$repo_name-$tag
eval "$(pixi shell-hook --manifest-path analysis)"
streamlit run TFinder-v1.py --server.port $PORT --server.headless true
Final directory structure:
── clock.time
├── img
├── LICENSE
├── navigation
├── README.md
├── requirements.txt
├── SECURITY.md
├── streamlit_modal
│ └── __init__.py
├── tests
│ ├── params.json
│ └── run_local.sh
├── tfinder
│ └── __init__.py
├── TFinder_health_universe.py
├── TFinder_standalone_win.bat
├── TFinder-v1.py
├── utils
│ └── components.py
├── river
│ └── main.sh
Step 5: Test Locally
bash tests/run_local.sh
The terminal will display access URLs:
Local URL: http://localhost:39905
Network URL: http://192.168.1.5:39905
External URL: http://1.54.25.47:39905
Step 6: Release
It has to add tag so the RIVER platform can fetch with specific version
git tag v1.0.0
git push origin main
git push origin v1.0.0